Allelic Imbalance
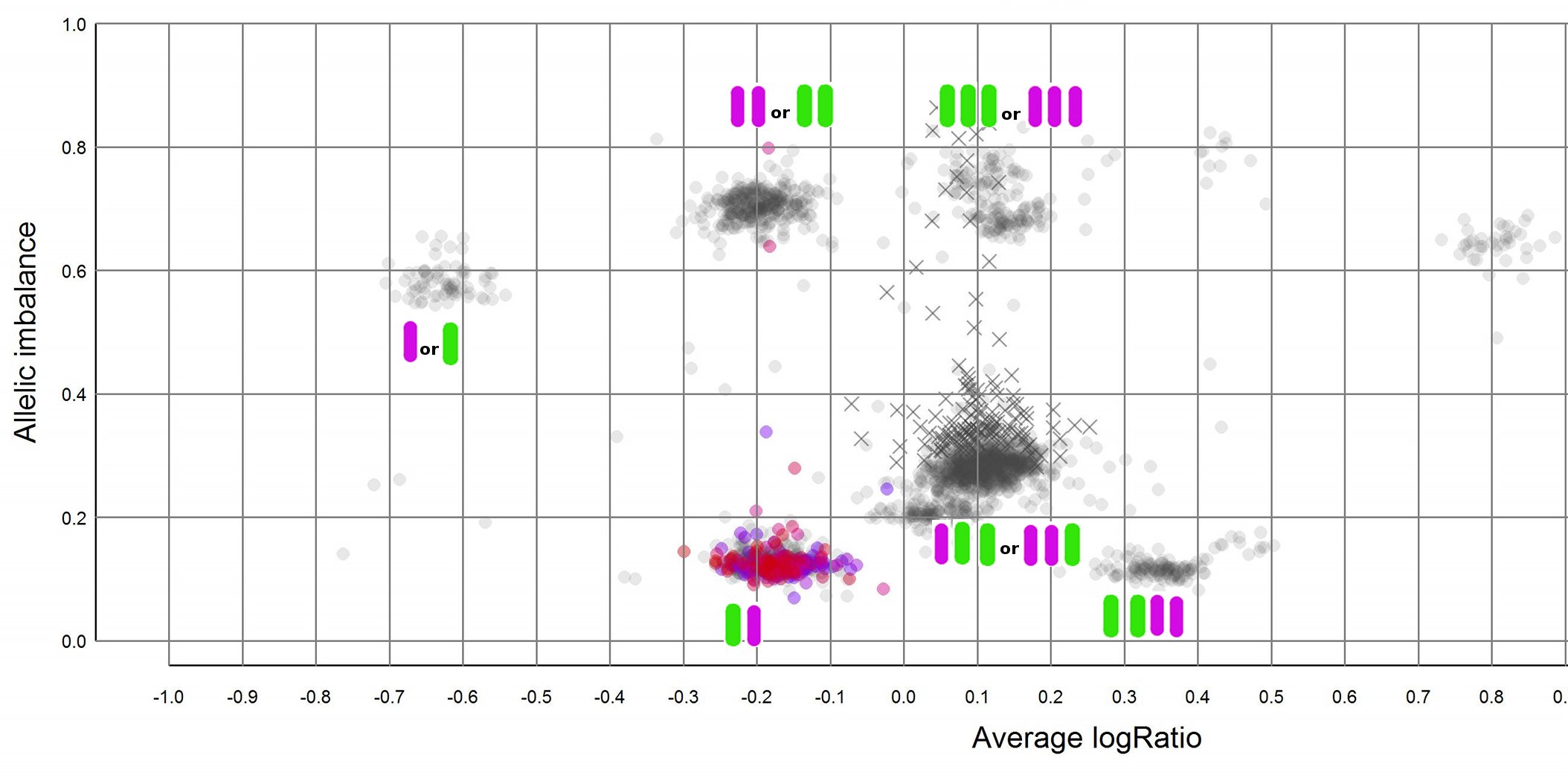
Allele-specific information is obtained from read counts at heterozygous SNPs. Allelic-imbalance separates chromosomal regions by their allele constitution. A diploid cell, which is the norm, will have one maternal and one paternal copy of a chromosome. For each copy number there can be losses of segments to either of the chromosomes as well as copies/gains. It is this allele-specific information that allelic imbalance captures. It does not, however, discern the origin of a segment, that is to say that two paternal copies have the same allelic imbalance ratio as two maternal copies. Below is an attempt to explain the different possible allelic combinations for a few copy numbers, it is VERY important that you understand that from an allelic imbalance value point of view it is the ratio between green and purple that is important, not if purple/green is maternal/paternal.


 is the same as
is the same as



 .
. | Copy Number | Possible chromosomal segment combinations | ||
|---|---|---|---|
| 1 |  |
||
| 2 | 
 |

 |
|
| 3 |


 |


 |
|
| 4 | 


 |



 |



 |
| 5 | 



 |




 |




 |
Here is a an example of a plot generated using TAPS where I have added the possible allele combinations next to the clusters.